
Overview
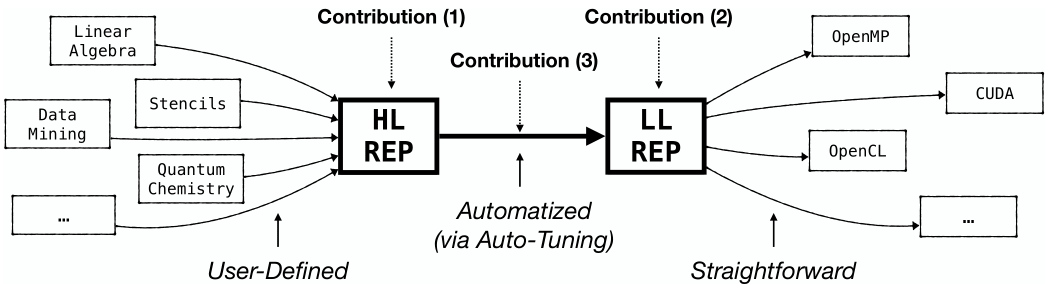
The approach of Multi-Dimensional Homomorphisms (MDH) is an algebraic formalism for systematically reasoning about de-composition and re-composition strategies of data-parallel computations (such as linear algebra routines and stencil computations) for the memory and core hierarchies of state-of-the-art parallel architectures (including GPUs, multi-core CPUs, as well as systems spanning multiple devices and nodes).
The MDH approach (formally) introduces:
- High-Level Program Representation (Contribution 1) that enables the user conveniently implementing data-parallel computations, agnostic from hardware and optimization details;
- Low-Level Program Representation (Contribution 2) that expresses device- and data-optimized de- and re-composition strategies of computations;
- Lowering Process (Contribution 3) that fully automatically lowers a data-parallel computation expressed in its high-level program representation to an optimized instance in its low-level representation, based on concepts from automatic performance optimization (a.k.a. auto-tuning), using the Auto-Tuning Framework (ATF).
The MDH’s low-level representation is designed such that Code Generation from it (e.g., in OpenMP for CPUs, CUDA for NVIDIA GPUs, or OpenCL for multiple kinds of architectures) becomes straightforward.
Our Experiments report encouraging results on GPUs and CPUs for MDH as compared to state-of-practice approaches, including NVIDIA cuBLAS/cuDNN and Intel oneMKL/oneDNN which are hand-optimized libraries provided by vendors.
Ultimate MDH Goals
- Performance: achieve performance competitive to hand-optimized solutions
- Portability: target any kind of parallel architecture
- Productivity: free user from hardware and optimization details
Getting Started
(Our implementation of MDH will be open sourced on GitHub)
Code Examples
From the following code examples, our MDH compiler fully automatically generates device- and data-optimized, executable program code, e.g., in OpenMP for CPUs, CUDA for NVIDIA GPUs, or OpenCL for multiple kinds of architectures.
MDH’s Python-Based User Interface
-
Matrix-Vector Multiplication (MatVec) expressed in MDH’s high-level program representation:
def matvec( T:BasicType, I:int,K:int ): @mdh() def mdh_matvec(): @scalar_function() def mul( out:Scalar[T], inp:Scalar[T,T] ): out['w'] = inp['M'] * inp['v'] @pw_custom_func() def add( res, lhs,rhs ): res['w'] = lhs['w'] + rhs['w'] return ( out_view[T]( w = [lambda i,k: (i)] ), md_hom[I,K]( mul, (cc,pw(add)) ), inp_view[T,T]( M = [lambda i,k: (i,k)] , v = [lambda i,k: (k) ] ) )The above defined
matvecfunction is used as follows:# MatVec on 1024x1024-sized input matrix and 1024-sized vector (both containing fp32 values) matvec__fp32__1024_1024 = matvec( fp32, 1024,1024 ) # CUDA host code ASSERT_DRV(cuda.cuInit(0)) device = ASSERT_DRV(cuda.cuDeviceGet(device_id)) # Get MDH "CUDA Module" for MatVec (using ATF-tuned optimizations) cuda__matvec__fp32__1024_1024 = mdhc.compile( computation=matvec__fp32__1024_1024, backend=CUDA( device ), tuner=pyATF( CUDARuntimeProfiler(), evaluations(1000) ) ) # MDH CUDA Module: compile CUDA code a100_cuda__matvec__fp32__1024_1024 = cuda__matvec__fp32__1024_1024.nvcc_compile( arch='compute_80' ) # MDH CUDA Module: run MatVec on M,v to obtain w w = np.random.rand(1024).astype(np.float32) M = np.random.rand(1024, 1024).astype(np.float32) v = np.random.rand(1024).astype(np.float32) a100_cuda__matvec__fp32__1024_1024.run( w,M,v ) # MDH CUDA Module: destroy module a100_cuda__matvec__fp32__1024_1024.destroy() -
Jacobi-1D (Jacobi1D) expressed in MDH’s high-level program representation:
def jacobi1d( T:BasicType, I:int ): @mdh() def mdh_jacobi1d(): @scalar_function() def f_j1d( out:Scalar[T], inp:Scalar[T,T,T] ): out['y'] = (inp['x', 1] + inp['x', 2] + inp['x', 3]) / 3.0 return ( out_view[T]( y = [lambda i: (i)] ), md_hom[I]( f_j1d, (cc) ), inp_view[T]( x = [lambda i: (i + 0), lambda i: (i + 1), lambda i: (i + 2)] ) )The above defined
jacobi1dfunction is used as follows:# Jacobi1D on a 1024-sized input vector (containing fp32 values) jacobi1d__fp32_1024 = jacobi1d( fp32, 1024 ) # CUDA host code ASSERT_DRV(cuda.cuInit(0)) device = ASSERT_DRV(cuda.cuDeviceGet(device_id)) # Get MDH "CUDA Module" for Jacobi1D (using ATF-tuned optimizations) cuda__jacobi1d__fp32_1024 = mdhc.compile( computation=jacobi1d__fp32_1024, backend=CUDA( device ), tuner=pyATF( CUDARuntimeProfiler(), evaluations(1000) ) ) # MDH CUDA Module: compile CUDA code a100_cuda__jacobi1d__fp32_1024 = cuda__jacobi1d__fp32_1024.nvcc_compile( arch='compute_80' ) # MDH CUDA Module: run Jacobi1D on x to obtain y y = np.random.rand(1024).astype(np.float32) x = np.random.rand(1026).astype(np.float32) a100_cuda__jacobi1d__fp32_1024.run( y,x ) # MDH CUDA Module: destroy module a100_cuda__jacobi1d__fp32_1024.destroy()
MDH’s MLIR-Based User Interface
-
func.func @main() { %M = memref.alloc() : memref<128x64xf32> %v = memref.alloc() : memref<64xf32> %w = mdh.compute "mdh_matvec" { inp_view = [ [ affine_map<( i,k ) -> ( i,k )> ], [ affine_map<( i,k ) -> ( k ) > ] ], md_hom = { scalar_func = @mul, combine_ops = [ "cc", ["pw",@add] ] }, out_view = [ [ affine_map<( i,k ) -> ( i )> ] ] } { inp_types = [ f32, f32 ], mda_size = [ 128,64 ], out_types = [ f32 ] }( %M,%v ) : ( memref<128x64xf32> , memref<64xf32> ) -> memref<128xf32> return }Here, functions
@muland@addare straightforward, user-defined functions for computing scalar multiplication or scalar addition, respectively (both not shown for brevity). Functionsccandpware pre-implemented combine operators for computing concatenation (cc) or point-wise operations (pw), respectively. -
func.func @main() { %x = memref.alloc() : memref<64xf32> %y = mdh.compute "mdh_jacobi1d" { inp_view = [ [ affine_map<( i ) -> ( i+0 )> , affine_map<( i ) -> ( i+1 )> , affine_map<( i ) -> ( i+2 )> ] ], md_hom = { scalar_func = @jacobi, combine_ops = [ "cc" ] }, out_view = [ [ affine_map<( i ) -> ( i )> ] ] } { inp_types = [ f32 ], mda_size = [ 62 ], out_types = [ f32 ] }( %x ) : (memref<64xf32>) -> (memref<62xf32>) return }Here, function
@jacobiis the Jacobi-specific computation (not shown for brevity), andccis a pre-implemented combine operator for computing concatenation.
Automatic Parallelization & Optimization
Additionally, MDH supports as inputs – as an alternative to DSL programs in MDH’s high-level programming interface (shown above) – also straightforward (annotated) sequential program code. For our MatVec example, our Python-based input code is of the following form:
def matvec( T:BasicType, I:int,K:int ):
@mdh( out( w = Buffer[T] ) ,
inp( M = Buffer[T], v = Buffer[T] ) ,
combine_ops( cc, pw(add) ) )
def mdh_matvec( w, M,v ):
for i in range(I):
for k in range(K):
w[i] = M[i,k] * v[k]
This program is completely equivalent to the DSL-based MDH program for MatVec shown above and used exactly the same:
# MatVec on 1024x1024-sized input matrix and 1024-sized vector (both containing fp32 values)
matvec__fp32__1024_1024 = matvec( fp32, 1024,1024 )
# CUDA host code
ASSERT_DRV(cuda.cuInit(0))
device = ASSERT_DRV(cuda.cuDeviceGet(device_id))
# Get MDH "CUDA Module" for MatVec (using ATF-tuned optimizations)
cuda__matvec__fp32__1024_1024 = mdhc.compile(
computation=matvec__fp32__1024_1024,
backend=CUDA( device ),
tuner=pyATF( CUDARuntimeProfiler(), evaluations(1000) )
)
# MDH CUDA Module: compile CUDA code
a100_cuda__matvec__fp32__1024_1024 = cuda__matvec__fp32__1024_1024.nvcc_compile( arch='compute_80' )
# MDH CUDA Module: run MatVec on M,v to obtain w
w = np.random.rand(1024).astype(np.float32)
M = np.random.rand(1024, 1024).astype(np.float32)
v = np.random.rand(1024).astype(np.float32)
a100_cuda__matvec__fp32__1024_1024.run( w,M,v )
# MDH CUDA Module: destroy module
a100_cuda__matvec__fp32__1024_1024.destroy()
Schedule-Based Optimization Process
MDH optionally allows expert users to incorporate knowledge into the optimization process using its scheduling language. By incorporating the user into the optimization process, we enable two major advantages over the standard MDH workflow:
- better optimization, as an auto-tuning system might not always make the same high-quality optimization decisions as a human expert
- faster auto-tuning, as some (or even all) optimization decisions might be made by the expert user and thus are not left to the costly auto-tuner
(An example scheduling program follows soon)
Publications
-
A. Rasch
(De/Re)-Composition of Data-Parallel Computations via Multi-Dimensional Homomorphisms
ACM Transactions on Programming Languages and Systems (TOPLAS 2024) – Presented at PLDI’24 conference
Paper Slides -
A. Rasch
Full Version: (De/Re)-Composition of Data-Parallel Computations via Multi-Dimensional Homomorphisms
arXiv 2024
Paper Slides -
A. Rasch, R. Schulze, D. Shabalin, A. Elster, S. Gorlatch, M. Hall
(De/Re)-Compositions Expressed Systematically via MDH-Based Schedules
ACM SIGPLAN International Conference on Compiler Construction (CC 2023)
Paper Slides
Talks
-
A. Rasch, R. Schulze
Code Generation & Optimization for Data-Parallel Computations via MDH and ATF
NextSilicon Seminar Series 2025; online event
Slides -
A. Rasch, R. Schulze
(De/Re)-Composition of Array Computations via Multi-Dimensional Homomorphisms + An MDH-Based DSL for Array Computations
ACM SIGPLAN Conference on Programming Language Design and Implementation (PLDI 2025); Seoul, Korea
Slides Talk -
A. Rasch, R. Schulze
MDH+ATF: Code Generation & Optimization for Deep-Learning Computations on NVIDIA GPUs
NVIDIA Codegen Community Forum 2025; online event
Slides -
A. Rasch, R. Schulze
MDH+ATF: Code Generation & Optimization for Deep-Learning Computations
Roofline AI 2025; Cologne, Germany
Slides -
A. Rasch, R. Schulze
MDH+ATF: Code Generation & Optimization for Deep-Learning Computations
Compilers for Machine Learning (C4ML 2025); Las Vegas NV, USA
Slides -
A. Rasch
Toward Performance & Portability & Productivity in Parallel Programming
The International Conference for High Performance Computing, Networking, Storage, and Analysis (SC 2024); Atlanta GA, USA
Slides -
A. Rasch
(De/Re)-Composition of Data-Parallel Computations via Multi-Dimensional Homomorphisms
ACM SIGPLAN Conference on Programming Language Design and Implementation (PLDI 2024); Copenhagen, Denmark
Slides -
Jens Hunloh, Lars Hunloh, R. Schulze, A. Rasch, T. Grosser
Linalg vs MDH: A Comparison of two MLIR Dialects
European LLVM Developers’ Meeting (EuroLLVM 2024); Vienna, Austria
Slides -
A. Rasch, R. Schulze, Jens Hunloh, Lars Hunloh
Code Generation & Optimization for Deep-Learning Computations via Multi-Dimensional Homomorphisms
Compilers for Machine Learning (C4ML 2024), (lightning talk)
Slides 1. Poster 2. Poster -
A. Rasch, R. Schulze, S. Gorlatch
Array Programming via Multi-Dimensional Homomorphisms
ACM SIGPLAN Conference on Programming Language Design and Implementation (PLDI 2023), (WIP paper)
Paper Slides Talk -
R. Schulze, A. Rasch, S. Gorlatch
Code Generation & Optimization for Deep-Learning Computations on GPUs via Multi-Dimensional Homomorphisms
International Conference for High Performance Computing, Networking, Storage and Analysis (SC 2021), Best Poster Finalist, (short paper)
Paper Poster Slides Talk -
A. Rasch, R. Schulze, S. Gorlatch
Using MLIR for Multi-Dimensional Homomorphisms
Google SIG MLIR Open Design Meeting 2020, (invited talk)
Slides Talk
Citations
Please use the following citations, when referring to MDH’s:
- Formalism & Design
@article{10.1145/3665643, author = {Rasch, Ari}, title = {(De/Re)-Composition of Data-Parallel Computations via Multi-Dimensional Homomorphisms}, year = {2024}, publisher = {Association for Computing Machinery}, address = {New York, NY, USA}, issn = {0164-0925}, url = {https://doi.org/10.1145/3665643}, doi = {10.1145/3665643}, journal = {ACM Trans. Program. Lang. Syst.}, month = {may}} - Scheduling Language
@inproceedings{10.1145/3578360.3580269, author = {Rasch, Ari and Schulze, Richard and Shabalin, Denys and Elster, Anne and Gorlatch, Sergei and Hall, Mary}, title = {(De/Re)-Compositions Expressed Systematically via MDH-Based Schedules}, year = {2023}, isbn = {9798400700880}, publisher = {Association for Computing Machinery}, address = {New York, NY, USA}, url = {https://doi.org/10.1145/3578360.3580269}, doi = {10.1145/3578360.3580269}, booktitle = {Proceedings of the 32nd ACM SIGPLAN International Conference on Compiler Construction}, pages = {61-72}, numpages = {12}, keywords = {GPU, scheduling languages, deep learning, CPU}, location = {Montr\'{e}al, QC, Canada}, series = {CC 2023} } - Automatic Parallelization and Optimization
@INPROCEEDINGS{mdpoly, author={Rasch, Ari and Schulze, Richard and Gorlatch, Sergei}, booktitle={Proceedings of the International Workshop on Polyhedral Compilation Techniques (IMPACT 2020)}, title={\texttt{md\_poly}: A Performance-Portable Polyhedral Compiler based on Multi-Dimensional Homomorphisms}, year={2020}, volume={}, number={}, pages={1-4}}
Contact

Ari Rasch
| Focus: | Project Lead |
| Affiliation: | University of Münster |
| Email: | a.rasch@uni-muenster.de |
| Website: | arirasch.net |

Richard Schulze
| Focus: | Technical Lead |
| Affiliation: | University of Münster |
| Email: | r.schulze@uni-muenster.de |
| Website: | richardschulze.net |
You can also find us on 

